Takanari Inoue, Ph.D.
Department of Cell Biology
Johns Hopkins University School of Medicine
855 N. Wolfe Street, 476 Rangos
Baltimore, MD 21205
Academic Titles
Professor of Cell Biology; Director, Center for Cell DynamicsResearch Topic
Synthetic cell biology, driven by hypothesis, technology and curiosity
One of the major challenges in cell biology is to understand how biochemical reactions at the molecular scale self-organize to generate physiological forms and functions at the cellular and tissue scale. Although genetic and biochemical approaches have been invaluable in identifying key regulatory molecules, these classical approaches rarely have the temporal and spatial resolution required to reveal the molecular mechanisms underlying such self-organization in vivo. Our research aims to address this knowledge gap by developing innovative molecular actuators that can be used to perturb and assay molecular actions in live cells with high temporal and spatial precision. In the past decade, we have applied these tools to studies of innate immune functions such as chemotaxis, phagocytosis and degranulation, as well as subcellular organizations including primary cilia, microtubules and stress granules. Please see below for our recent achievements under this theme:
- Non-catalytic allostery in α-TAT1 by a phospho-switch drives dynamic microtubule acetylation
Journal of Cell Biology, 2022:221:11:e1-20
“Location, Location, Location”
We report an unconventional regulatory mechanism of α-TAT1, a posttranslational modification enzyme that acetylates microtubules. More specifically, we identified a unique signal motif in the intrinsically disordered region of α-TAT1. Multidisciplinary characterization subsequently revealed that this previously uncharacterized motif functions as an intricate molecular switch that integrates upstream kinase signaling to actuate the intracellular localization of α-TAT1, hence achieving dynamic microtubule acetylation. Activity of cellular enzymes is often regulated allosterically, where the interaction between a given enzyme and its substrate is initiated by a third molecule binding to the enzyme to induce conformational change. In contrast, it appears to be the spatial regulation that mainly controls the functional output of α-TAT1. While there are enzymes that translocate between nucleus and cytosol, α-TAT1 is the only enzyme to our knowledge that uses the nucleus as a sequestration hideout.
In parallel to characterizing α-TAT1 regulatory mechanisms, we developed a co-recruitment assay based on chemically inducible dimerization tools to assess protein-protein interactions. This assay is like performing Co-IP assays in living cells, and is quantitative, robust and reliable. Most existing protein-protein interaction assays require purification of proteins and/or lysing cells where procedural concerns need to be taken into consideration for data interpretation. Our co-recruitment assays circumvent this problem. And thanks to the modular design principles, the assay is generalizable to virtually any soluble proteins of your choice. Therefore, we expect its wide application to diverse systems.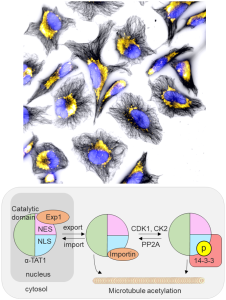
2. A molecular trap inside microtubules probes luminal access by soluble proteins
Nature Chemical Biology, 2021:17:888-895
“Illuminating the path to cellular bower”
We present the first method to chemically and inducibly trap proteins inside microtubules that enables direct assessment of biomolecular accessibility in living cells. This work overcame previous technical limitations and provides a novel mechanistic insight of how protein regulators and effectors gain access to the site of their action inside the cellular tunnel.
Microtubules are the fundamental basis for many cellular functions including cell division, cell migration, neuronal plasticity and ciliogenesis. The multi-tasking schemes of microtubule functions is believed to be encoded in a spatio-temporal pattern as well as a combinatorial set of their posttranslational modification status, a concept put forth as the tubulin code. While these modifications mostly occur on the outer surface, microtubules can also be regulated via the inner part, such as via acetylation. As a cellular tunnel, the microtubules uniquely present a lumen that is physically insulated by the cylindrical wall of densely-packed tubulins, with limited connections to the cytoplasm at their open ends. It is natural to wonder how regulatory and effector proteins of acetylated tubulins reach this secluded site.
The bottleneck of microtubule studies thus far is the limited number of experimental systems. To overcome this challenge, we rationally established a chemically-inducible technique for conditional trapping of a cytosolic protein in the microtubule lumen, with which we subsequently discovered that soluble proteins can enter the lumen via diffusion through openings at the MT ends and sides. Additionally, proteins forming a complex with tubulins can be incorporated to the lumen through the plus ends. We believe that our molecular tool is unique and powerful in probing biomolecular accessibility, with a strong potential in decoding tubulin codes in the near future, thus appealing ubiquitously to a large audience in cell biology.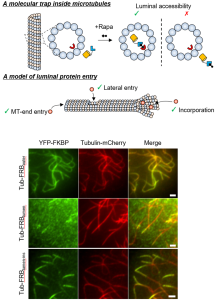
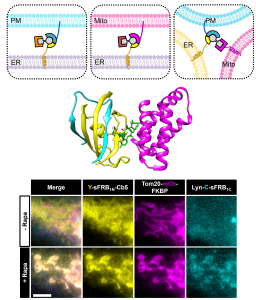
3. Rational design and implementation of a chemically inducible hetero-trimerization system
Nature Methods, 2020:17:928–936
“The Three Musketeers”
We report the first chemically inducible “trimerization” system (CIT), where a small molecule rapidly brings three unique proteins together in living cells, offering unprecedented intracellular operation in addressing new biological questions, harnessing cell functions and behaviors, as well as performing synthetic operations.
Since the development of a chemically inducible dimerization system (CID) in the late 90’s, a number of CID tools were generated and used to advance life sciences in answering biological questions. In addition, the CID-mediated operation became a foundation for designing optogenetic dimerization tools in recent years. Despite the prevalent trimerization events in nature (death receptors inducing apoptosis, G proteins initiating signal transduction, etc.), and despite the self-evident utility of CIT, we do not have CIT tools yet in our hands; this is simply because developing CIT is extremely challenging.
In this study, our multidisciplinary team constructed a trimerization system by co-opting unique properties of the well-established FKBP/FRB/rapamycin dimerization system, thus rationally avoiding the challenging task of engineering multiple cooperative protein-small molecule interactions de novo. Through computational analysis, we identified split sites for the FKBP or FRB proteins such that the two split protein halves and the cognate protein partner form the three stable protein components of the trimerization system. Live-cell imaging experiments and structural validation with protein crystallography showed that the split-based CIT system undergoes rapid trimerization upon rapamycin addition. Increased capability in spatiotemporal control conferred by CIT allowed us to demonstrate its potential, for example, in the emerging field of organelle-organelle membrane contact sites. Since CIT molecular tools are modular in design, these first CIT tools pave the way for future generalizable CIT-based approaches that alter function at organelle junctions and offer higher spatiotemporal control in other biological contexts. The work was highlighted in Nature Chemical Biology, “Three’s company”.